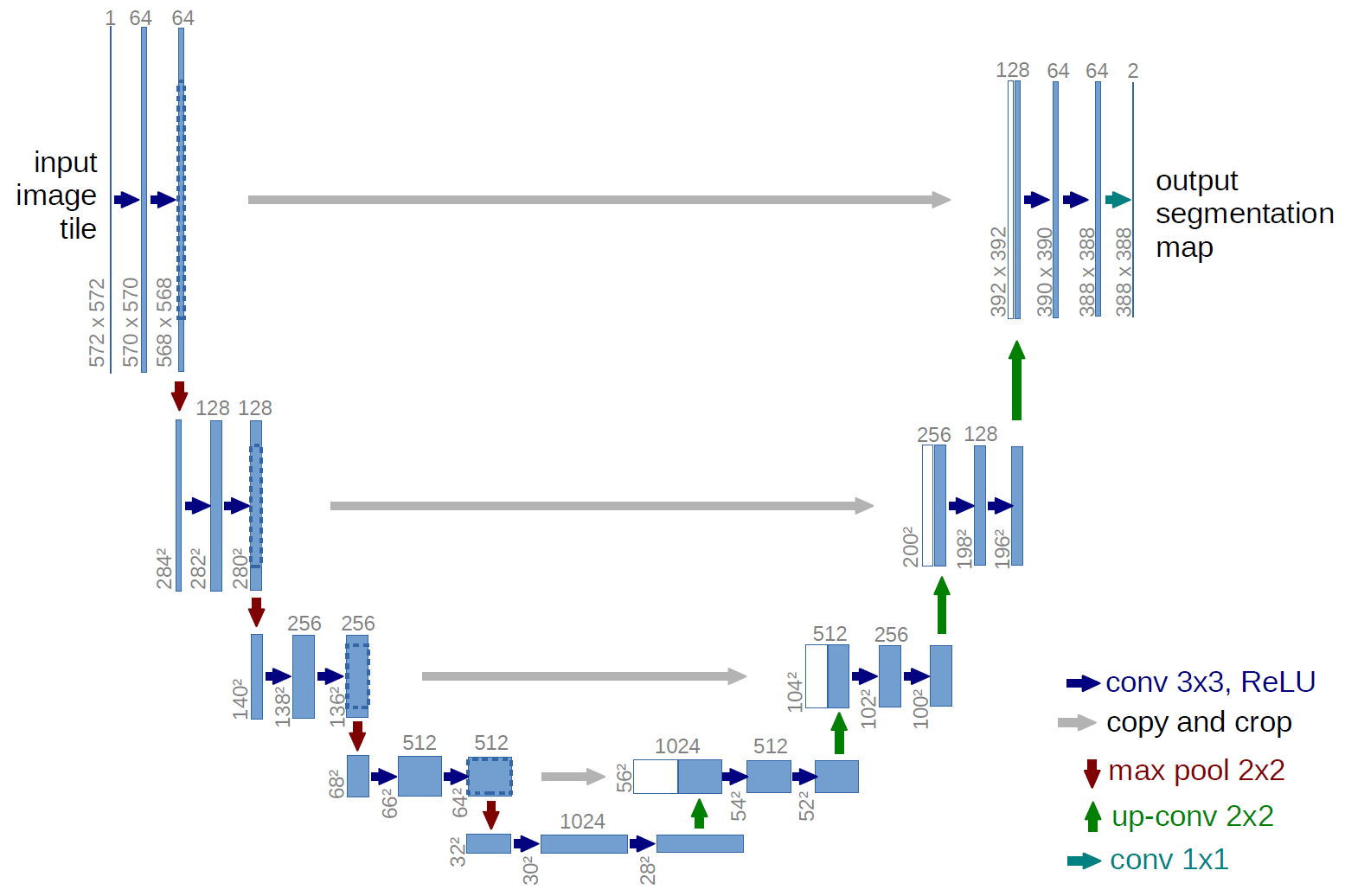
U-Net Architecture
What is U-Net?
- U-Net is a convolutional neural network (CNN) architecture designed for biomedical image segmentation.
- It was introduced by Olaf Ronneberger, Philipp Fischer, and Thomas Brox in their 2015 paper titled “U-Net: Convolutional Networks for Biomedical Image Segmentation.” The architecture is named “U-Net” because of its U-shaped structure.
Key Features
1. Encoder(Contracting Path) :Captures context by progressively reducing the spatial dimensions while increasing the number of feature channels.Important features of encoders :
Feature Extraction: Extracts hierarchical features, from low-level edges to high-level structures.
Dimensionality Reduction: Reduces spatial dimensions to focus on important features and reduce computation.
Contextual Understanding: Captures global context for better understanding of the image.
Building the Feature Hierarchy: Creates a multi-level feature abstraction for the decoder to use.
Transfer Learning Compatibility: Can be pre-trained on large datasets and fine-tuned for specific tasks.
Bottleneck Layer: Summarizes the input into a compressed, abstract representation.
Adaptability to Different Input Sizes: Handles varying input sizes due to flexible convolutional operations.
Modularity: Can be replaced or modified with other architectures for improved performance.
2. Decoder(Expansive Path):Recovers spatial dimensions through up sampling and combines features from the encoder to localize the segmentation.Important features of decoders:
Feature Reconstruction: Reconstructs the spatial dimensions of the image from the encoder’s compressed features.
Upsampling: Increases spatial resolution to restore the original image size.
Skip Connections Integration: Combines fine-grained details from the encoder for precise localization.
Segmentation Map Generation: Produces the final output (e.g., pixel-wise segmentation map).
Contextual Refinement: Refines global context from the encoder with local details for accurate predictions.
Hierarchical Feature Fusion: Merges multi-level features from the encoder for better feature representation.
End-to-End Learning: Trains alongside the encoder to optimize the entire network for the task.
Flexibility: Can adapt to different output resolutions based on the task requirements.
Modularity: Can be customized or replaced with other upsampling or decoding architectures.
3. Skip Connections : These connections link corresponding layers in the encoder and decoder, allowing the network to retain fine-grained spatial information, which is crucial for precise segmentation.
- Directly transfer fine-grained spatial details from the encoder to the decoder, enabling precise localization and improving segmentation accuracy by combining high-level context with low-level features.
Variants of U-Net
| Variants | Description | Applications |
|---|---|---|
| Attention U-Net | Adds attention mechanisms to the U-Net architecture. Attention gates (AGs) focus on the most relevant regions of the input while ignoring irrelevant areas. | Medical imaging, remote sensing |
| 3D U-Net | Extends the U-Net to 3D by replacing 2D convolutional layers with 3D convolutions. It processes volumetric data like 3D medical scans. | Volumetric data (MRI, CT scans) |
| Residual U-Net | Incorporates residual connections (borrowed from ResNet) within U-Net. These connections help in training deeper networks by addressing vanishing gradient issues. | High-resolution or complex boundary segmentation |
| Dense U-Net | Combines the U-Net with DenseNet principles, where each layer is connected to every other layer in the same block. | Retinal vessel segmentation, rich feature extraction |
| U-Net++ (Nested) | Improves upon U-Net by introducing a nested architecture with multiple intermediate U-Nets. It uses dense skip connections to aggregate features across different scales. | Fine-grained segmentation, remote sensing |
| R2U-Net | Combines residual and recurrent connections into U-Net to model sequential dependencies and capture long-term contextual information. | Video segmentation, sequential data |
| Attention ResU-Net | Merges the concepts of attention gates, residual connections, and U-Net to emphasize relevant features while maintaining efficient gradient flow. | Histopathology, cardiac imaging |
| Lightweight U-Net | A smaller version of U-Net designed for resource-constrained environments. It uses depthwise separable convolutions and fewer parameters. | Mobile applications, real-time segmentation |
| U-Net3+ | Enhances U-Net++ by introducing deeper skip pathways and full-scale feature fusion. It focuses on multi-scale feature learning. | Multi-scale object segmentation |
| TransUNet | Combines U-Net with transformer-based architectures to capture both local and global dependencies in images. | Large-scale image segmentation, medical imaging |
| UNet-GAN | Combines U-Net with a GAN framework, using U-Net as the generator and a discriminator to evaluate segmentation quality. | Synthetic masks, retinal image segmentation |
| SegNet | A similar encoder-decoder structure as U-Net but uses max-pooling indices to improve the upsampling process. | Road and scene segmentation |
| Bayesian U-Net | Extends U-Net with Bayesian modeling to estimate uncertainty in predictions. | Uncertainty estimation, risk-aware applications |
| Multi-Scale U-Net | Incorporates multi-scale feature extraction techniques to improve segmentation across objects of varying sizes. | Satellite imagery, large variation segmentation |
Why popular now?
U-Net, despite being introduced in 2015, has been gaining renewed attention in recent years because of its versatility, adaptability, and increasing relevance in cutting-edge AI domains.
1. Growth of Diffusion Models :
U-Net plays a crucial role in diffusion models, especially in denoising diffusion probabilistic models (DDPMs) and stable diffusion, due to its ability to effectively handle high-dimensional data and extract meaningful features.
| Applications | How U-Net Contributes |
|---|---|
| Text-to-Image Generation | In models like Stable Diffusion, U-Net integrates text embeddings (via cross-attention) and performs latent space denoising for high-quality image synthesis. |
| Image-Super-resolution | U-Net handles noise removal and detail enhancement to upscale low-resolution images effectively. |
| Inpainting | U-Net reconstructs missing regions of an image by utilizing its encoder-decoder structure to learn contextual and spatial details. |
| Image-to-Image Translation | U-Net takes a noisy or degraded input and translates it into the desired output (e.g., style transfer, segmentation, or restoration tasks). |
| Video Synthesis | U-Net is extended to 3D for processing spatiotemporal data in video diffusion models. |
| Medical Imaging | Diffusion models with U-Net enhance and denoise medical images for better diagnostics or analysis. |
2. Medical Imaging Boom
U-Net was originally designed for bio-medical image segmentation, and this field has expanded significantly due to advancements in imaging technologies and AI-powered diagnostics.
With increasing demand for accurate segmentation of organs, tumors, and lesions in applications like:
Cancer detection (lung, brain, breast, etc.).
Automated surgical assistance.
Personalized treatment planning.
U-Net remains the gold standard for these tasks, and its variants (e.g., 3D U-Net, Attention U-Net) continue to drive innovation.
3. Foundation for Transformer-Hybrid Architectures
Recent architectures like TransUNet and U-Net with attention mechanisms combine the strengths of U-Net’s hierarchical processing with transformer-based global attention mechanisms. Applications includes Text-to-image generation (e.g., Stable Diffusion), Medical imaging (leveraging both local and global context) etc.
Code from Scratch
import torch
import torch.nn as nn
# Define a basic convolutional block
# Performs two sets of Conv2d → BatchNorm → ReLU operations.
# Encapsulates the basic building block used in the encoder, decoder, and bottleneck.
class ConvBlock(nn.Module):
def __init__(self, in_channels, out_channels):
super(ConvBlock, self).__init__()
self.conv = nn.Sequential(
nn.Conv2d(in_channels, out_channels, kernel_size=3, padding=1),
nn.BatchNorm2d(out_channels),
nn.ReLU(inplace=True),
nn.Conv2d(out_channels, out_channels, kernel_size=3, padding=1),
nn.BatchNorm2d(out_channels),
nn.ReLU(inplace=True),
)
def forward(self, x):
return self.conv(x)
# Define the encoder block
# Contains a ConvBlock followed by a MaxPool2d layer for downsampling.
# Outputs both the feature map (used for skip connections) and the downsampled output.
class EncoderBlock(nn.Module):
def __init__(self, in_channels, out_channels):
super(EncoderBlock, self).__init__()
self.conv = ConvBlock(in_channels, out_channels)
self.pool = nn.MaxPool2d(kernel_size=2, stride=2)
def forward(self, x):
x = self.conv(x)
x_down = self.pool(x)
return x, x_down
# Define the decoder block
# Upsamples the input using ConvTranspose2d.
# Crops and concatenates the corresponding encoder feature map (skip connection).
# Applies a ConvBlock to refine the features.
class DecoderBlock(nn.Module):
def __init__(self, in_channels, out_channels):
super(DecoderBlock, self).__init__()
self.upconv = nn.ConvTranspose2d(in_channels, out_channels, kernel_size=2, stride=2)
self.conv = ConvBlock(in_channels, out_channels)
def forward(self, x, skip_connection):
x = self.upconv(x)
# Ensure the dimensions match due to cropping (for odd-sized inputs)
diffY = skip_connection.size(2) - x.size(2)
diffX = skip_connection.size(3) - x.size(3)
x = nn.functional.pad(x, [diffX // 2, diffX - diffX // 2, diffY // 2, diffY - diffY // 2])
x = torch.cat((skip_connection, x), dim=1)
return self.conv(x)
# U-Net architecture
# Encoder Path: Iteratively downsamples input while extracting features.
# Bottleneck: Processes the smallest-scale features.
# Decoder Path: Upsamples and refines features, using skip connections.
# Final Layer: Outputs the segmentation map via a 1x1 convolution.
class UNet(nn.Module):
def __init__(self, in_channels=1, out_channels=1, features=[64, 128, 256, 512]):
super(UNet, self).__init__()
self.encoder_blocks = nn.ModuleList()
self.decoder_blocks = nn.ModuleList()
self.bottleneck = ConvBlock(features[-1], features[-1] * 2)
# Encoder
for feature in features:
self.encoder_blocks.append(EncoderBlock(in_channels, feature))
in_channels = feature
# Decoder
for feature in reversed(features):
self.decoder_blocks.append(DecoderBlock(feature * 2, feature))
self.final_conv = nn.Conv2d(features[0], out_channels, kernel_size=1)
def forward(self, x):
skip_connections = []
# Encoder path
for encoder in self.encoder_blocks:
x, x_down = encoder(x)
skip_connections.append(x)
x = x_down
# Bottleneck
x = self.bottleneck(x)
# Decoder path
skip_connections = skip_connections[::-1]
for idx, decoder in enumerate(self.decoder_blocks):
x = decoder(x, skip_connections[idx])
return self.final_conv(x)
# Instantiate the model and print summary
# Input: (Batch, Channels, Height, Width), e.g., (1, 1, 256, 256).
# Output: Same spatial dimensions as the input for segmentation tasks.
if __name__ == "__main__":
model = UNet(in_channels=1, out_channels=1) # For grayscale input and binary segmentation
x = torch.randn((1, 1, 256, 256)) # Batch size = 1, 1 channel, 256x256 image
preds = model(x)
print(f"Input shape: {x.shape}")
print(f"Output shape: {preds.shape}")